Note
Go to the end to download the full example code.
Reaction Equation#
The reaction equation is probably the simples non-trivial unsteady differenital equation. It is given by equation (1). If the forcing \(f\) is only space dependent, then it has a very simple analytical solution given by (2), where \(u_\mathrm{initial} = u(x, y, 0)\) and \(u_\mathrm{final} = f(x, y) / \alpha\).
In the 2D case with differential geometry, this problem can be solved either by taking \(u\) to be a 0-form or a 2-form. For this example, the 0-form is taken.
For the error measurements used to check for convergence, both \(L^2\) and \(H^1\) norms will be used to compute error for each time step. These values will then be integrated using trapezoidal integration to find total error over the entire time march. This error will then be compared for different number of time steps over the same time period.
import numpy as np
import numpy.typing as npt
import pyvista as pv
import rmsh
from matplotlib import pyplot as plt
from mfv2d import (
ConvergenceSettings,
KFormSystem,
KFormUnknown,
SolverSettings,
SystemSettings,
TimeSettings,
UnknownFormOrder,
mesh_create,
solve_system_2d,
)
from scipy.integrate import trapezoid
Setup#
The initial and final solutions were taken as \(u_\mathrm{initial} = 2 \cos(\frac{\pi x}{2}) \cos(\frac{\pi y}{2})\) and \(u_\mathrm{final} = \sin(\pi x) \cos(\pi y)\). As for the value of the rate coefficient, the value of \(\alpha = 0.25\) was chosen.
The problem would be simulated on the time interval \(t \in [0, 10]\).
T_END = 10
ALPHA = 0.25
def initial_u(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Screw initial solution."""
return 2 * np.cos(np.pi * x / 2) * np.cos(np.pi * y / 2)
def initial_q(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Screw initial curl."""
return np.stack(
(
-np.pi * np.cos(np.pi * x / 2) * np.sin(np.pi * y / 2),
np.pi * np.sin(np.pi * x / 2) * np.cos(np.pi * y / 2),
),
axis=-1,
)
def final_u(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Steady state forcing."""
return np.sin(np.pi * x) * np.cos(np.pi * y)
def final_q(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Steady state curl."""
return np.stack(
(
-np.pi * np.sin(np.pi * x) * np.sin(np.pi * y),
-np.pi * np.cos(np.pi * x) * np.cos(np.pi * y),
),
axis=-1,
)
System Setup#
The system being solved is formulated bellow. An additional equation was introducted to obtain the curl of the solution, so that the \(H^1\) norm could be computed.
u = KFormUnknown("u", UnknownFormOrder.FORM_ORDER_0)
v = u.weight
q = KFormUnknown("q", UnknownFormOrder.FORM_ORDER_1)
p = q.weight
system = KFormSystem(
ALPHA * (v @ u) == ALPHA * (v @ final_u),
p @ q - p @ u.derivative == 0,
sorting=lambda f: f.order,
)
Make the Mesh#
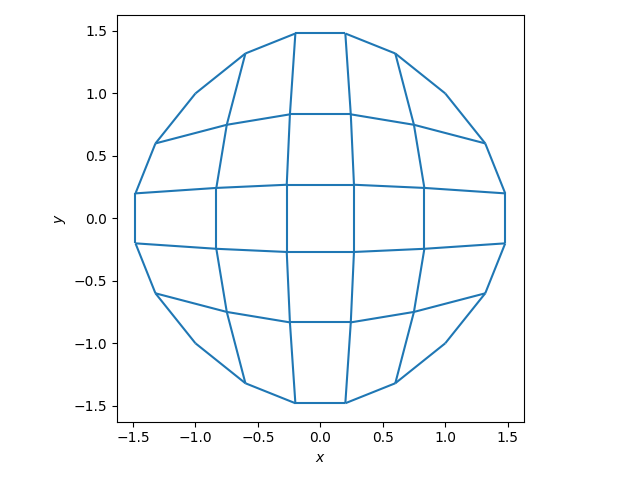
Next the mesh would be created. In this case, it was taken to be a convexly deformed square.
N = 6
P = 4
n1 = N
n2 = N
rect_mesh, rx, ry = rmsh.create_elliptical_mesh(
rmsh.MeshBlock(
label=None,
bottom=rmsh.BoundaryCurve.from_knots(n1, (-1, -1), (0, -2), (+1, -1)),
right=rmsh.BoundaryCurve.from_knots(n2, (+1, -1), (+2, 0), (+1, +1)),
top=rmsh.BoundaryCurve.from_knots(n2, (+1, +1), (0, +2), (-1, +1)),
left=rmsh.BoundaryCurve.from_knots(n2, (-1, +1), (-2, 0), (-1, -1)),
)
)
assert rx < 1e-6 and ry < 1e-6
mesh = mesh_create(
P,
np.stack((rect_mesh.pos_x, rect_mesh.pos_y), axis=-1),
rect_mesh.lines + 1,
rect_mesh.surfaces,
)
fig, ax = plt.subplots(1, 1)
xlim, ylim = rect_mesh.plot(ax)
ax.set(
aspect="equal",
xlim=(1.1 * xlim[0], 1.1 * xlim[1]),
ylim=(1.1 * ylim[0], 1.1 * ylim[1]),
xlabel="$x$",
ylabel="$y$",
)
fig.tight_layout()
plt.show()
Run Unsteady Simulations#
With the mesh and system defined, the simulations can be run. The run is done for 10, 20, 50, and 100 time steps.
nt_vals = np.array((10, 20, 50, 100))
h1_err = np.zeros(nt_vals.size)
l2_err = np.zeros(nt_vals.size)
dt_vals = np.zeros(nt_vals.size)
for i_nt, nt in enumerate(nt_vals):
dt = float(T_END / nt)
solutions, stats, mesh = solve_system_2d(
mesh,
system_settings=SystemSettings(system, initial_conditions={u: initial_u}),
solver_settings=SolverSettings(
ConvergenceSettings(
maximum_iterations=10, relative_tolerance=0, absolute_tolerance=1e-10
)
),
time_settings=TimeSettings(dt=dt, nt=nt, time_march_relations={v: u}),
recon_order=10,
)
n_sol = len(solutions)
h1_err_vals = np.zeros(n_sol)
l2_err_vals = np.zeros(n_sol)
time_vals = np.zeros(n_sol)
for isol, sol in enumerate(solutions):
time = float(sol.field_data["time"][0])
u_exact = initial_u(sol.points[:, 0], sol.points[:, 1]) * np.exp(
-ALPHA * time
) + final_u(sol.points[:, 0], sol.points[:, 1]) * (1 - np.exp(-ALPHA * time))
q_exact = initial_q(sol.points[:, 0], sol.points[:, 1]) * np.exp(
-ALPHA * time
) + final_q(sol.points[:, 0], sol.points[:, 1]) * (1 - np.exp(-ALPHA * time))
u_err = sol.point_data["u"] - u_exact
q_err = sol.point_data["q"] - q_exact
sol.point_data["u_err"] = (u_err) ** 2
# sol.point_data["u_real"] = u_exact
sol.point_data["q_err"] = np.linalg.norm(q_err, axis=-1)
# sol.point_data["q_real"] = q_exact
integrated = sol.integrate_data()
time_vals[isol] = time
h1_err_vals[isol] = integrated.point_data["q_err"][0]
l2_err_vals[isol] = np.sqrt(integrated.point_data["u_err"][0])
total_h1_error = trapezoid(h1_err_vals, time_vals)
total_l2_error = trapezoid(l2_err_vals, time_vals)
h1_err[i_nt] = total_h1_error
l2_err[i_nt] = total_l2_error
dt_vals[i_nt] = dt
# print(f"For {dt=} total error was {total_h1_error:.3e}.")
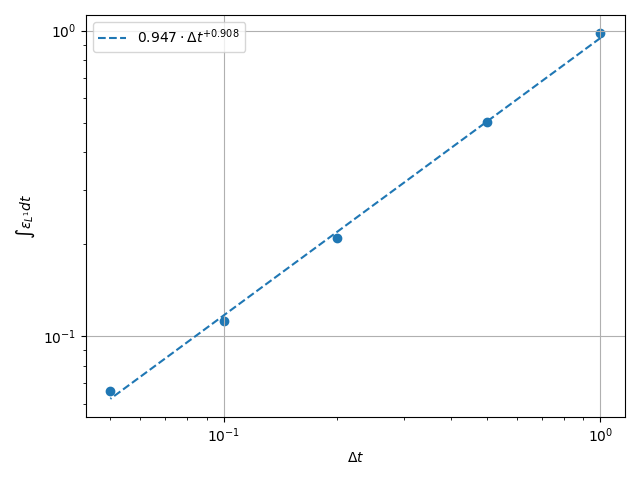
Plot the Time Error#
The total integrated time error in the two norms is now examined.
\(H^1\) Norm#
k1, k0 = np.polyfit(np.log(dt_vals), np.log(h1_err), 1)
k0 = np.exp(k0)
fig, ax = plt.subplots(1, 1)
ax.scatter(dt_vals, h1_err)
ax.plot(
dt_vals,
k0 * dt_vals**k1,
linestyle="dashed",
label=f"${k0:.3g} \\cdot {{\\Delta t}}^{{{k1:+.3g}}}$",
)
ax.grid()
ax.legend()
ax.set(
xlabel="$\\Delta t$",
ylabel="$\\int \\varepsilon_{H^{1}} {dt}$",
xscale="log",
yscale="log",
)
ax.xaxis_inverted()
fig.tight_layout()
plt.show()

\(L^2\) Norm#
k1, k0 = np.polyfit(np.log(dt_vals), np.log(l2_err), 1)
k0 = np.exp(k0)
fig, ax = plt.subplots(1, 1)
ax.scatter(dt_vals, l2_err)
ax.plot(
dt_vals,
k0 * dt_vals**k1,
linestyle="dashed",
label=f"${k0:.3g} \\cdot {{\\Delta t}}^{{{k1:+.3g}}}$",
)
ax.grid()
ax.legend()
ax.set(
xlabel="$\\Delta t$",
ylabel="$\\int \\varepsilon_{L^{1}} {dt}$",
xscale="log",
yscale="log",
)
ax.xaxis_inverted()
fig.tight_layout()
plt.show()
Plot Solution’s Evolution#
With pyvista the unsteady solution can even be plotted.
plotter = pv.Plotter(off_screen=True, window_size=(1600, 800))
plotter.open_gif("unsteady-reaction-direct-solution.gif", fps=30)
for sol in solutions:
sol.points[:, 2] = sol.point_data["u"]
plotter.add_mesh(sol, scalars=None, name="solution", show_scalar_bar=False)
plotter.add_text(f"time = {sol.field_data['time'][0]:.1f}", name="time")
plotter.write_frame()
plotter.close()
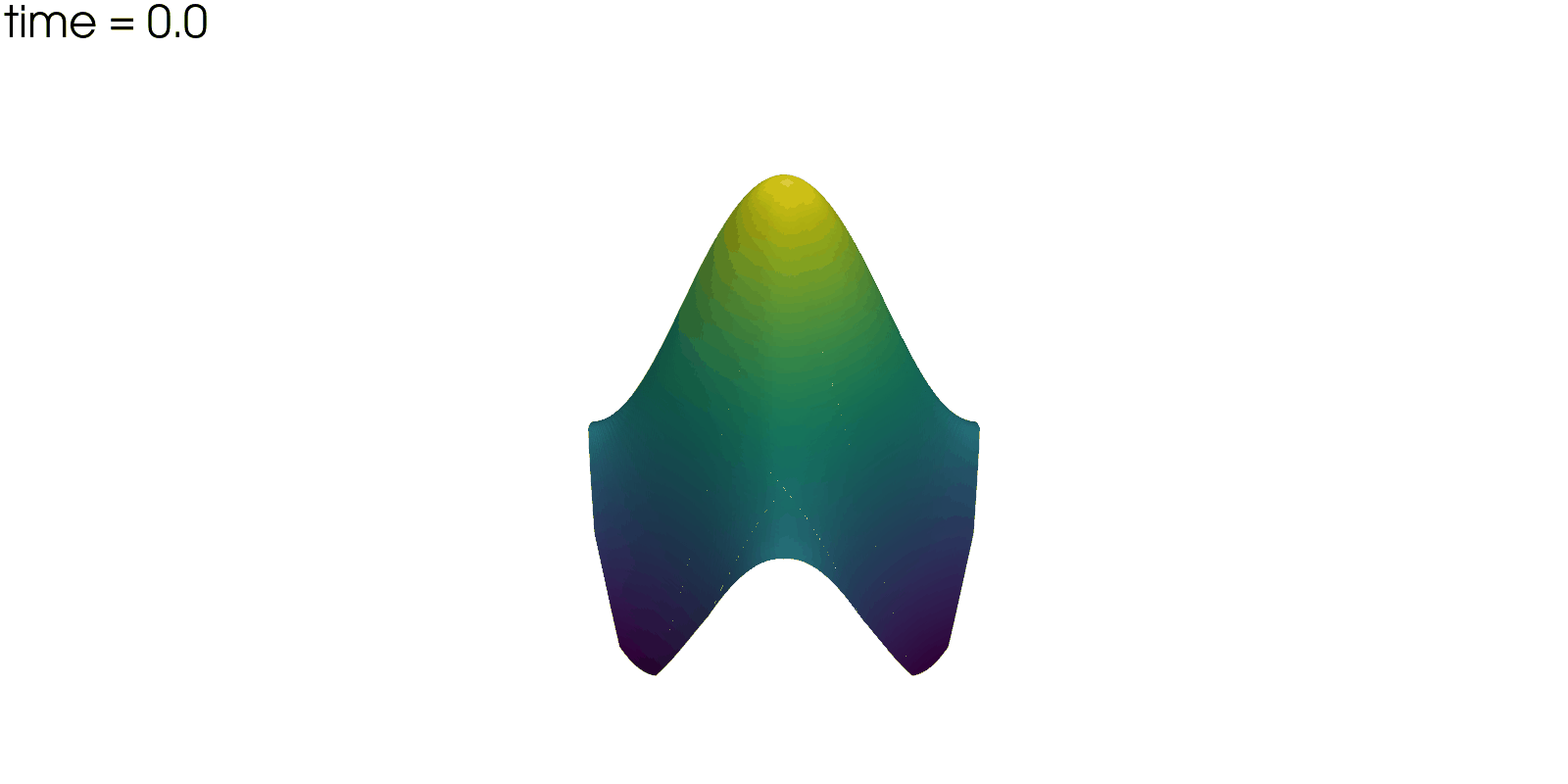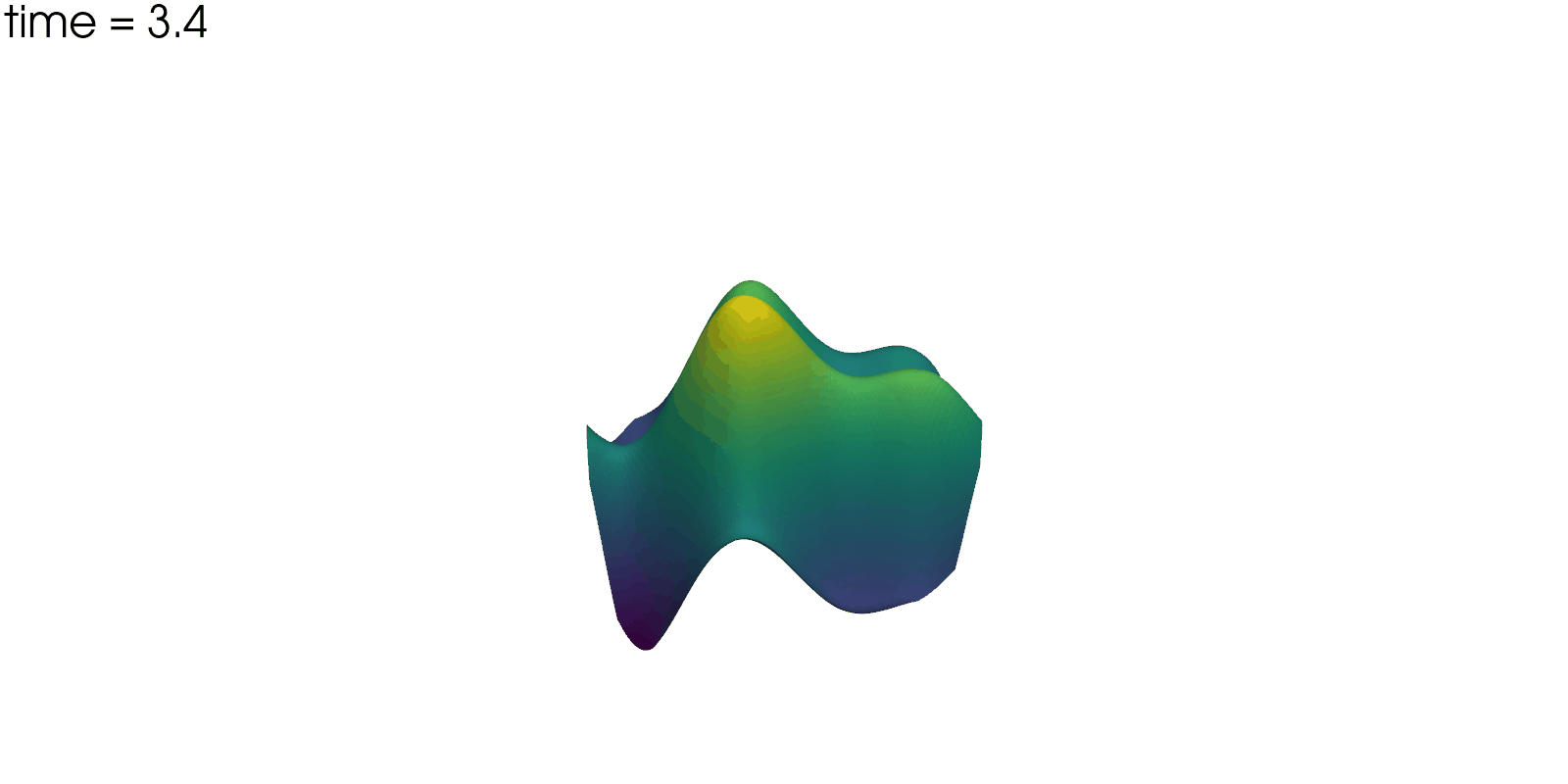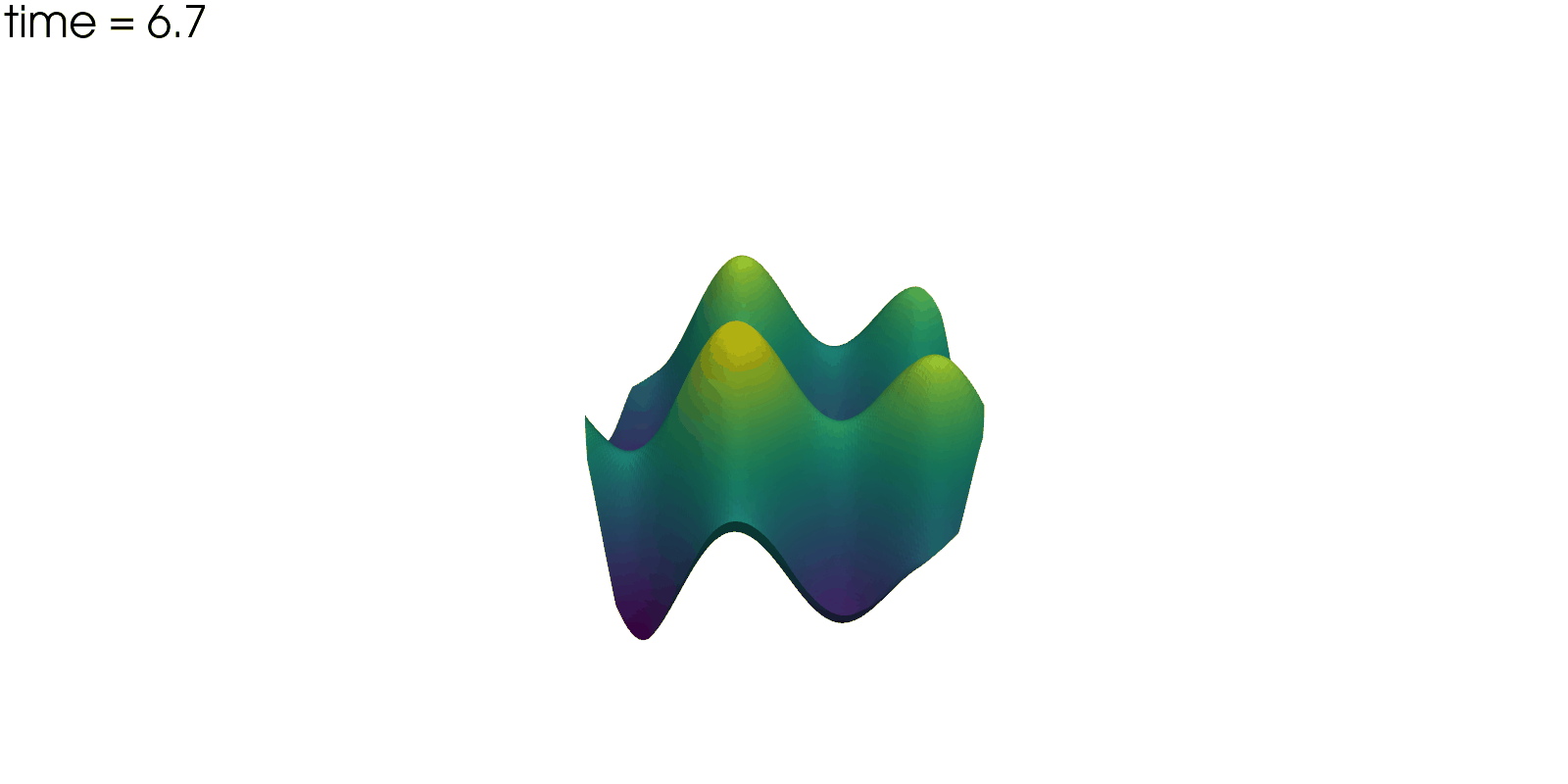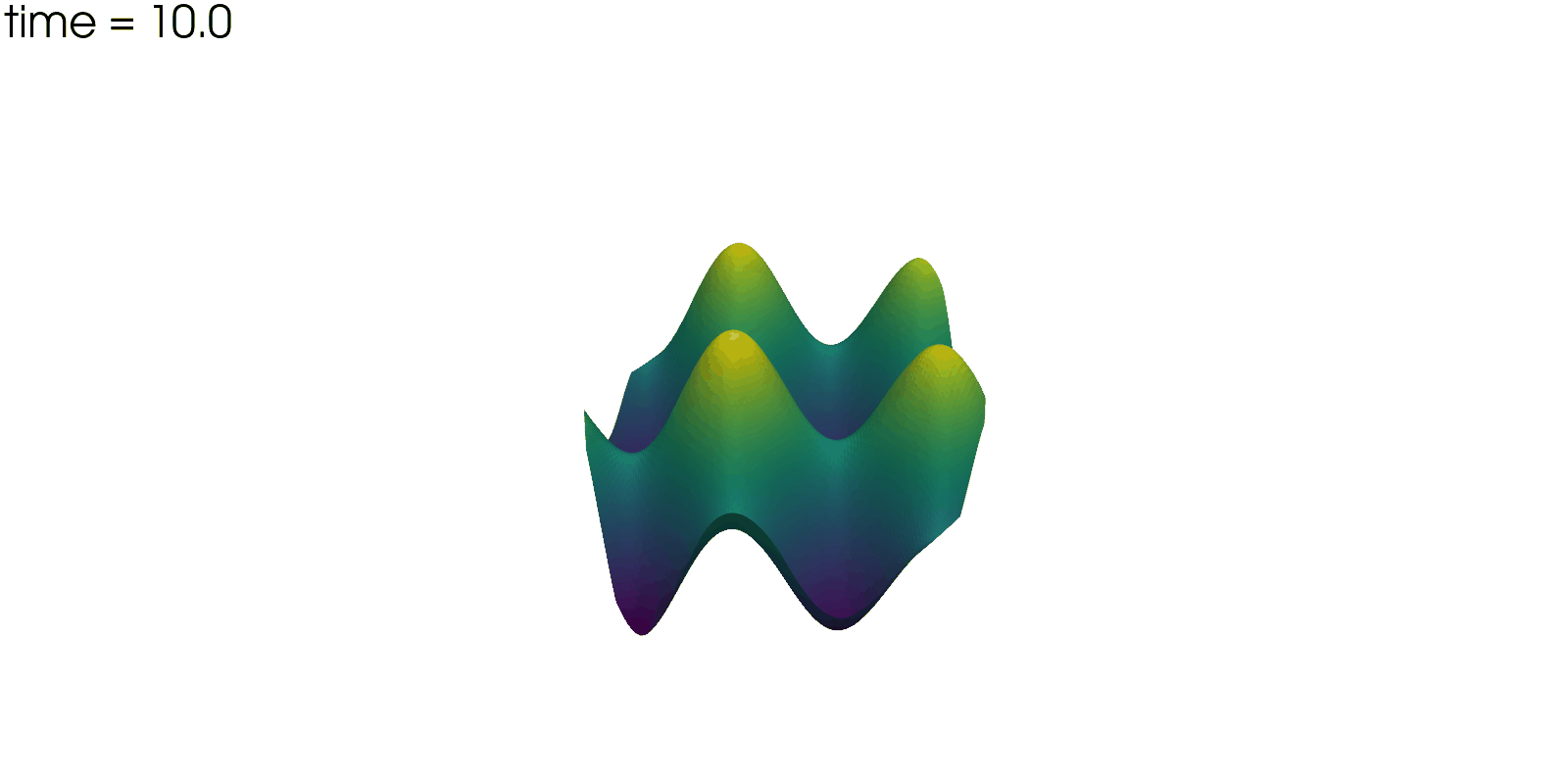
Total running time of the script: (0 minutes 32.835 seconds)