Note
Go to the end to download the full example code.
Unsteady Heat Equations in Mixed Formulation#
This example is exactly the same as Unsteady Heat Equations in Direct Formulation. As such, only differences from that one will be mentioned.
import numpy as np
import numpy.typing as npt
import rmsh
from matplotlib import pyplot as plt
from mfv2d import (
ConvergenceSettings,
KFormSystem,
KFormUnknown,
SolverSettings,
SystemSettings,
TimeSettings,
UnknownFormOrder,
mesh_create,
solve_system_2d,
system_as_string,
)
from scipy.integrate import trapezoid
Setup#
Since the mixed formulation naturally also gives the graident, it is also computed here.
ALPHA = 0.02
BETA = 1
def steady_u(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Steady state solution."""
return np.cos(np.pi * x / 2) * np.cos(np.pi * y / 2)
def steady_q(x: npt.NDArray[np.floating], y: npt.NDArray[np.floating]):
"""Steady state gradient."""
return np.stack(
(
-np.pi / 2 * np.sin(np.pi * x / 2) * np.cos(np.pi * y / 2),
-np.pi / 2 * np.cos(np.pi * x / 2) * np.sin(np.pi * y / 2),
),
axis=-1,
)
u = KFormUnknown("u", UnknownFormOrder.FORM_ORDER_2)
v = u.weight
q = KFormUnknown("q", UnknownFormOrder.FORM_ORDER_1)
p = q.weight
system = KFormSystem(
p.derivative @ u - p @ q == p ^ steady_u,
ALPHA * (v @ q.derivative)
== BETA * (v @ steady_u) - (BETA - ALPHA * np.pi**2 / 2) * (v @ u),
sorting=lambda f: f.order,
)
print(system_as_string(system))
N = 13
P = 3
T_END = 2
n1 = N
n2 = N
rect_mesh, rx, ry = rmsh.create_elliptical_mesh(
rmsh.MeshBlock(
label=None,
bottom=rmsh.BoundaryCurve.from_line(n1, (-1, -1), (+1, -1)),
right=rmsh.BoundaryCurve.from_line(n2, (+1, -1), (+1, +1)),
top=rmsh.BoundaryCurve.from_line(n2, (+1, +1), (-1, +1)),
left=rmsh.BoundaryCurve.from_line(n2, (-1, +1), (-1, -1)),
)
)
assert rx < 1e-6 and ry < 1e-6
mesh = mesh_create(
P,
np.stack((rect_mesh.pos_x, rect_mesh.pos_y), axis=-1),
rect_mesh.lines + 1,
rect_mesh.surfaces,
)
nt_vals = np.logspace(start=1, stop=5, num=6, base=2, dtype=np.uint32)
h1_err = np.zeros(nt_vals.size)
l2_err = np.zeros(nt_vals.size)
dt_vals = np.zeros(nt_vals.size)
for i_nt, nt in enumerate(nt_vals):
dt = float(T_END / nt)
solutions, stats, mesh = solve_system_2d(
mesh,
system_settings=SystemSettings(system),
solver_settings=SolverSettings(
ConvergenceSettings(
maximum_iterations=20, relative_tolerance=0, absolute_tolerance=1e-13
)
),
time_settings=TimeSettings(dt=dt, nt=nt, time_march_relations={v: u}),
recon_order=25,
)
n_sol = len(solutions)
h1_vals = np.zeros(n_sol)
l2_vals = np.zeros(n_sol)
time_vals = np.zeros(n_sol)
for isol, sol in enumerate(solutions):
time = float(sol.field_data["time"][0])
u_exact = steady_u(sol.points[:, 0], sol.points[:, 1]) * (
1 - np.exp(-BETA * time)
)
u_err = sol.point_data["u"] - u_exact
sol.point_data["u_err"] = u_err**2
sol.point_data["u_exact"] = u_exact
q_exact = steady_q(sol.points[:, 0], sol.points[:, 1]) * (
1 - np.exp(-BETA * time)
)
q_err = sol.point_data["q"] - q_exact
sol.point_data["q_err"] = np.linalg.norm(q_err, axis=-1)
sol.point_data["q_exact"] = q_exact
integrated = sol.integrate_data()
time_vals[isol] = time
h1_vals[isol] = integrated.point_data["q_err"][0]
l2_vals[isol] = np.sqrt(integrated.point_data["u_err"])[0]
# print(f"Error at time {time:.3g} is {err:.3e}")
h1_total_error = trapezoid(h1_vals, time_vals)
l2_total_error = trapezoid(l2_vals, time_vals)
h1_err[i_nt] = h1_total_error
l2_err[i_nt] = l2_total_error
dt_vals[i_nt] = dt
print(f"For {dt=:.3g} total error was {h1_total_error:.3e}.")
[-1.0 M(1) | (E(2, 1))^T M(2)] [q(1)] [+ B<q, steady_u>] [0 | 0 ] [q(1)]
[0.02 M(2) E(2, 1) | 0 ] [u(2)] = [+ E<u, steady_u>] + [0 | -0.9013039559891064 M(2)] [u(2)]
For dt=1 total error was 1.986e-01.
For dt=0.667 total error was 9.133e-02.
For dt=0.333 total error was 2.327e-02.
For dt=0.2 total error was 8.415e-03.
For dt=0.111 total error was 2.613e-03.
For dt=0.0625 total error was 8.627e-04.
Plotting the Error#
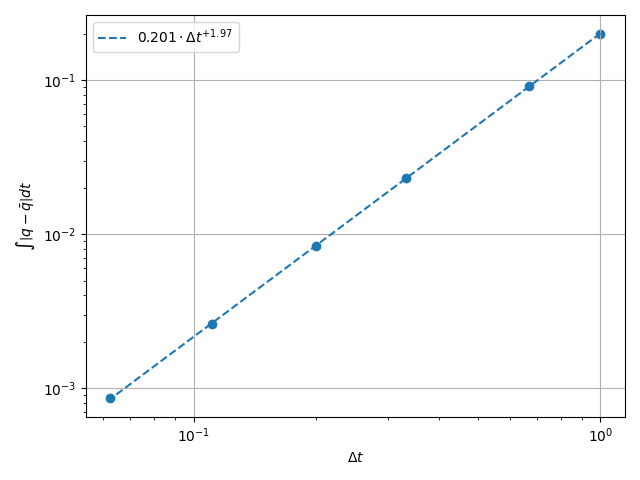
Now we plot the error. As you can see, we magically got another order of accuracy out of fucking thin air. If I had to guess it is related to the fact that the time integration is symplectic.
\(H^1\) Norm#
k1, k0 = np.polyfit(np.log(dt_vals), np.log(h1_err), 1)
k0 = np.exp(k0)
fig, ax = plt.subplots(1, 1)
ax.scatter(dt_vals, h1_err)
ax.plot(
dt_vals,
k0 * dt_vals**k1,
linestyle="dashed",
label=f"${k0:.3g} \\cdot {{\\Delta t}}^{{{k1:+.3g}}}$",
)
ax.grid()
ax.legend()
ax.set(
xlabel="$\\Delta t$",
ylabel="$\\int \\left|q - \\bar{q}\\right| {dt}$",
xscale="log",
yscale="log",
)
ax.xaxis_inverted()
fig.tight_layout()
plt.show()
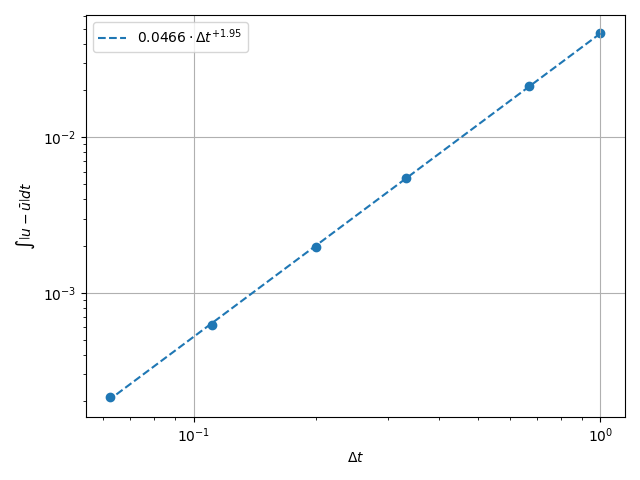
\(L^2\) Norm#
k1, k0 = np.polyfit(np.log(dt_vals), np.log(l2_err), 1)
k0 = np.exp(k0)
fig, ax = plt.subplots(1, 1)
ax.scatter(dt_vals, l2_err)
ax.plot(
dt_vals,
k0 * dt_vals**k1,
linestyle="dashed",
label=f"${k0:.3g} \\cdot {{\\Delta t}}^{{{k1:+.3g}}}$",
)
ax.grid()
ax.legend()
ax.set(
xlabel="$\\Delta t$",
ylabel="$\\int \\left|u - \\bar{u}\\right| {dt}$",
xscale="log",
yscale="log",
)
ax.xaxis_inverted()
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 10.991 seconds)